Key advances: global ocean viromes, host–virus metabolic coupling, discovery of new viral lineages
Prior to Tara Oceans, global knowledge of marine viral diversity and ecology was extremely limited. By combining targeted viral metagenomics with new analytical frameworks, Tara Oceans revealed that the oceans harbor hundreds of thousands of distinct viral populations, some of which represent entirely new phyla (Tara Oceans publication numbers 119, 129, 133, 141, 163). These efforts culminated in global ocean virome catalogs that fundamentally reshaped our understanding of viral diversity, host specificity, and biogeography (Tara Oceans publication numbers 23, 43, 72, 86, 129).
Analyses demonstrated that marine viruses actively reprogram host metabolism during infection through auxiliary metabolic genes (AMGs), influencing carbon, nitrogen, and sulfur cycling. Entirely new viral lineages and deep evolutionary connections between viruses and cellular life were uncovered (Tara Oceans publication numbers 43, 117, 126, 153, 163). Tara Oceans also provided the first global maps of eukaryotic viruses, including large DNA viruses (also known as NCLDVs), elucidating their environmental drivers and evolutionary roles, including impacts on host life cycles and population dynamics (Tara Oceans publication numbers 51, 92, 111, 161).
These discoveries established viruses as central regulators of marine ecosystems and key predictors of carbon export from surface waters to the deep ocean (Tara Oceans publication number 34, 108, 110).
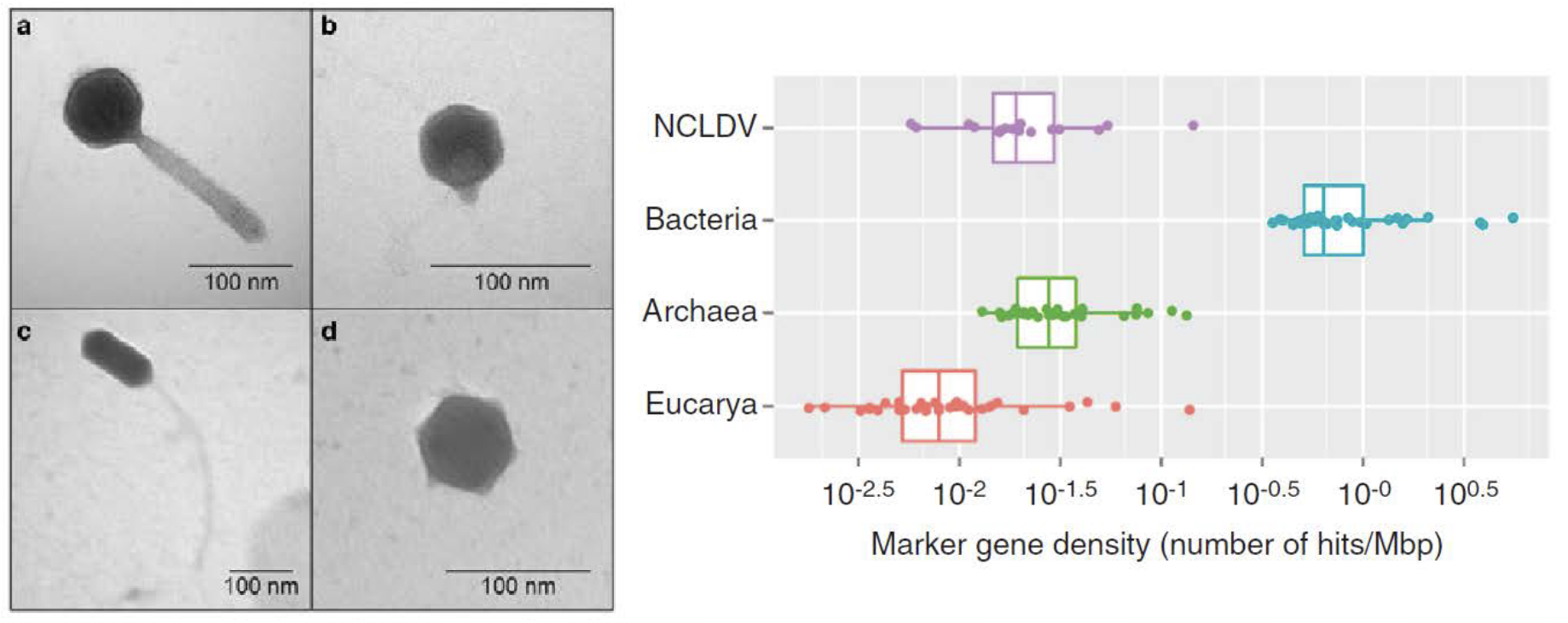
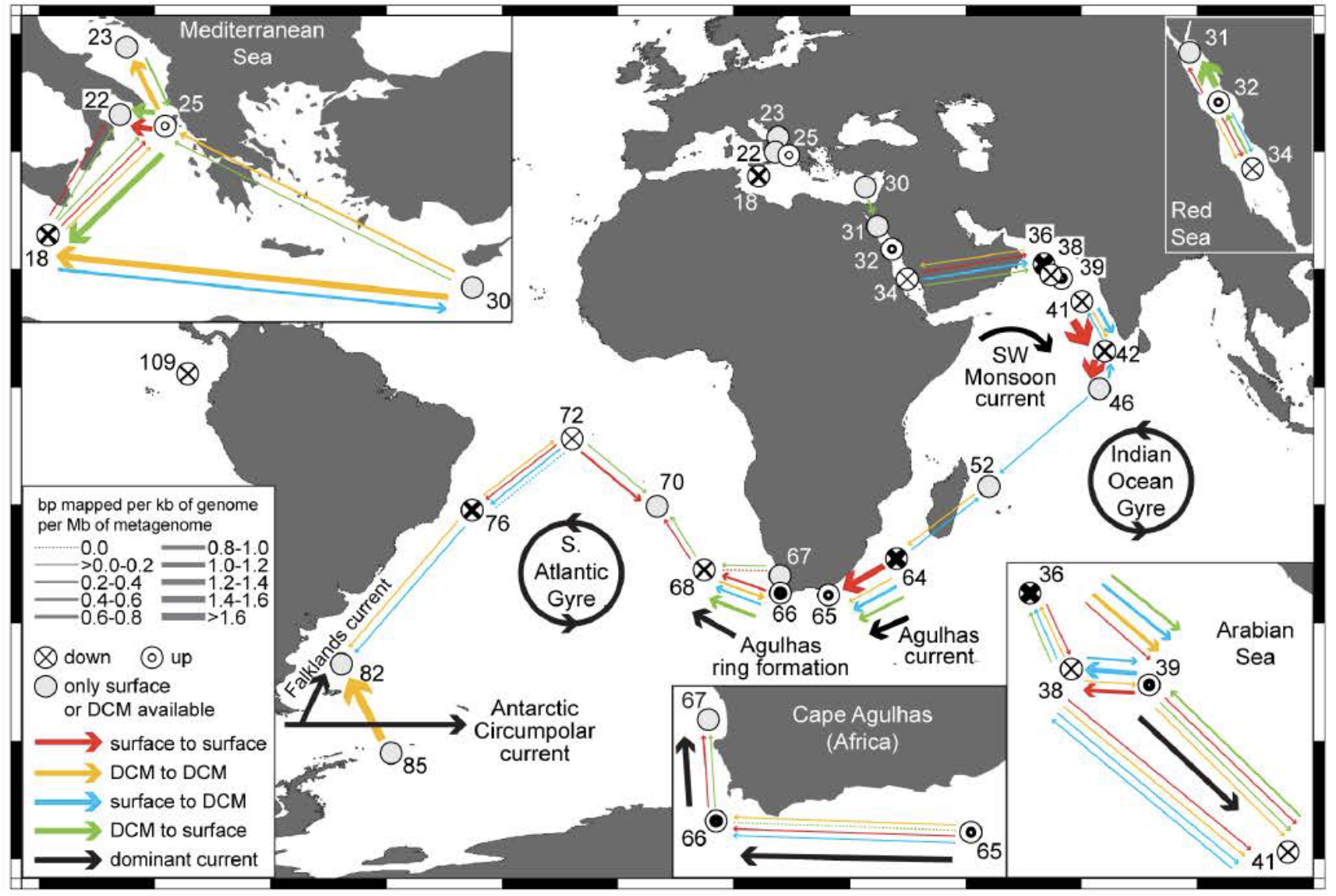
Figure 3 : Tara Oceans virus morpho-genetic study. Upper left: Morphologies of prokaryote- infecting viruses observed in Tara Oceans samples: (a) myovirus; (b) podovirus; (c) siphovirus; (d) non- tailed virus. The understudied non-tailed viruses emerged as being the most representative in our meta-omics datasets. Upper right: taxonomy marker genes suggest that the abundance of giant DNA viruses (NCLDV) infecting eukaryotes exceed the abundance of their host by one order of magnitude, a pattern reminiscent of the 1 to 10 ratio between the abundance of bacteria and phages. Bottom panel: (f) Our first global virome analysis indicated that viral communities were passively transported by oceanic currents and locally structured by environmental conditions that affect host community structure.
