Key advances: unprecedented eukaryotic diversity, parasitism and symbiosis, unified taxonomy
Protists represent the most diverse and complex component of marine plankton, spanning wide ranges of size, morphology, and trophic strategy. Tara Oceans produced the largest coherent morpho‑genetic dataset of planktonic eukaryotes ever assembled, integrating metabarcoding, metatranscriptomics, single‑cell genomics, and imaging across multiple size fractions.
Global analyses revealed extraordinary eukaryotic diversity, dominated by heterotrophic protists, parasites, and symbionts, many of which were previously unknown. Current estimates suggest 130,000 genetic types of protists, which is more than 10 times higher than the number of formally described species of marine eukaryotic plankton. Around one third cannot be assigned to any known taxonomic group (Tara Oceans publication number 24). Tara Oceans has also generated a collection of 116 million genes from marine eukaryotes – the largest catalogue of genes from a single biome (Tara Oceans publication number 62). Collectively, Tara Oceans data has transformed understanding of protist biogeography, trophic roles, and evolutionary history, while contributing to the development of a unified eukaryotic taxonomy through the UniEuk initiative (Tara Oceans publication numbers 37, 50, 68, 71, 76, 80, 89, 93, 125, 127, 156).
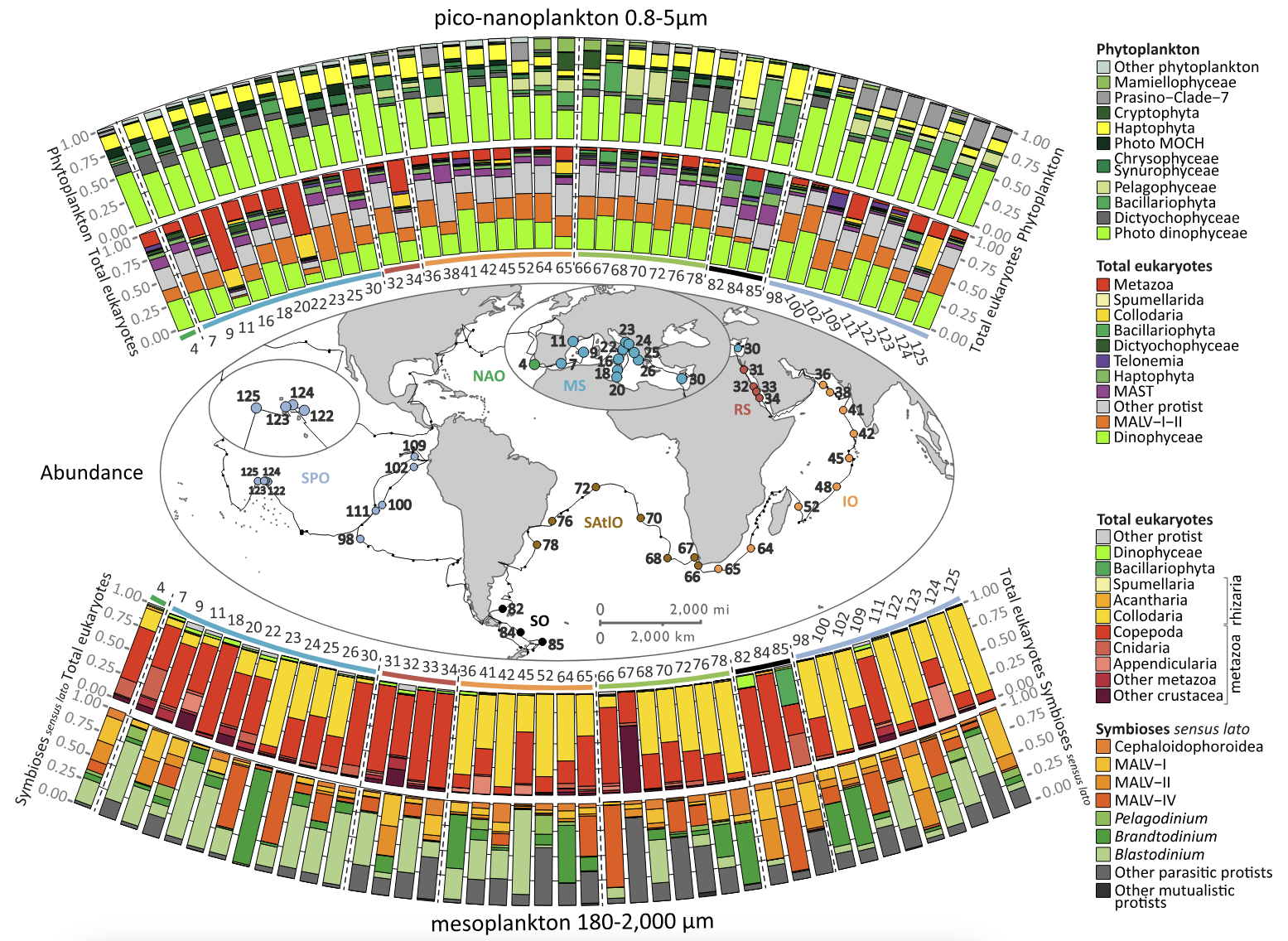
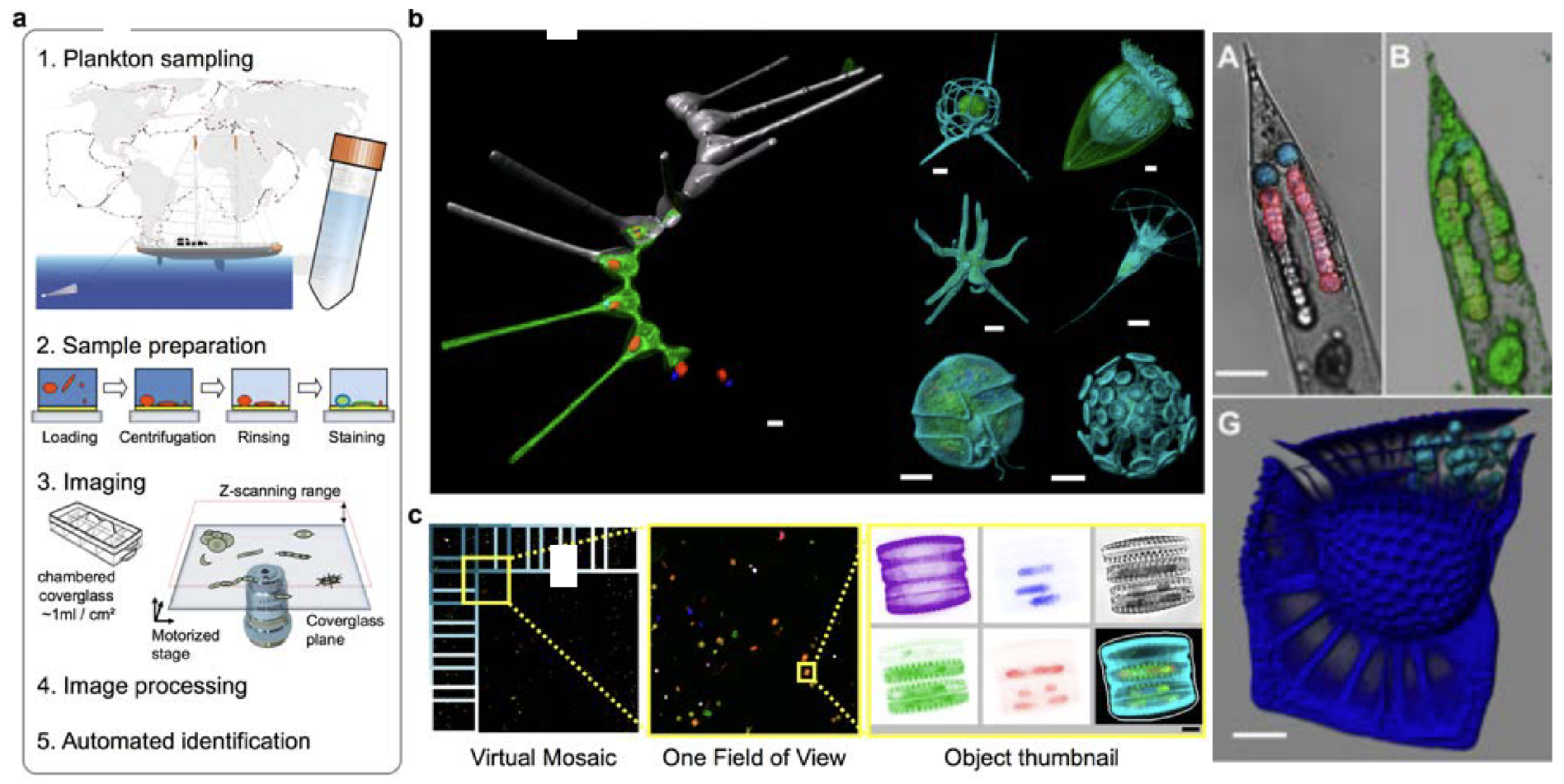
Figure 5. Upper panel: We used rDNA metabarcoding to estimate the relative biovolume of eukaryotic plankton taxa in the world sunlit ocean (above the map: phytoplankton and total eukaryotes in piconanoplankton; below the map: total eukaryotes and known symbiotic protists for the mesoplankton). The importance of dinophyceae and collodarians in the smallest and largest size- fractions, respectively, came as a surprise. Lower panel: For future, sub-cellular exploration of marine protists eco-biocomplexity, we developed eHCFM (environmental High Content Fluorescence Microscopy; Colin et al. eLIFE, in review). e-HCFM enables automated quantitative 3D-fluorescence imaging of eukaryotic cell structures (including symbionts; see for instance cyanobacterial symbionts in a diatom and a dinoflagellate on the right side) across the full diversity of microbial eukaryotes, while recognizing and classifying the imaged taxa.
