Key advances: baseline for marine ecosystems at the dawn of the 21st century, global plankton interactome, ecosystem‑level carbon export, circulation‑driven biogeography
By integrating viral, prokaryotic, eukaryotic, and imaging data, Tara Oceans enabled reconstruction of the first global plankton interactome, revealing dense networks of interactions across all domains of life (Tara Oceans publication numbers 25, 120). These analyses showed that biotic interactions often outweigh environmental factors in structuring plankton communities and identified parasites and symbionts as key connectors of food webs (Tara Oceans publication numbers 25, 70, 102). The Tara Oceans interactome infers that there are many acts of cooperation for every act of competition; that marine plankton ecosystems succeed because they consist of reciprocal interdependent systems of interactions between individuals in a range of symbiotic interactions whose fates are tied together. These findings invite us to reflect on the neodarwinian view of evolution that the most powerful driver of ecosystem organization is competition.
Approaches based on machine learning further linked plankton community structure and gene expression to carbon export, nutrient availability, ocean circulation and climate variability (Tara Oceans publication numbers 34, 93, 128, 146, 148), and genome-scale metabolic modeling is providing additional insights of relevance for understanding the ocean’s biogeochemical cycles (e.g., Tara Oceans publication number 150). Collectively, these studies established Tara Oceans as a reference framework for the global ocean in that it provides a baseline to describe the composition and functioning of marine ecosystems at the beginning of the 21st century. It’s highly standardized sampling and analysis pipelines permit systems-based approaches for exploring marine ecosystems and a critical foundation for predicting their responses to past and ongoing environmental change (Tara Oceans publication numbers 55, 80, 93, 128, 131).
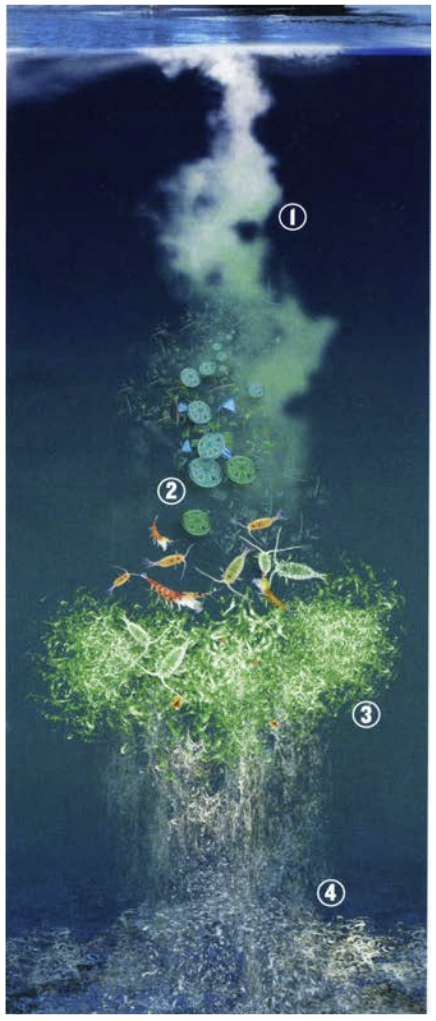
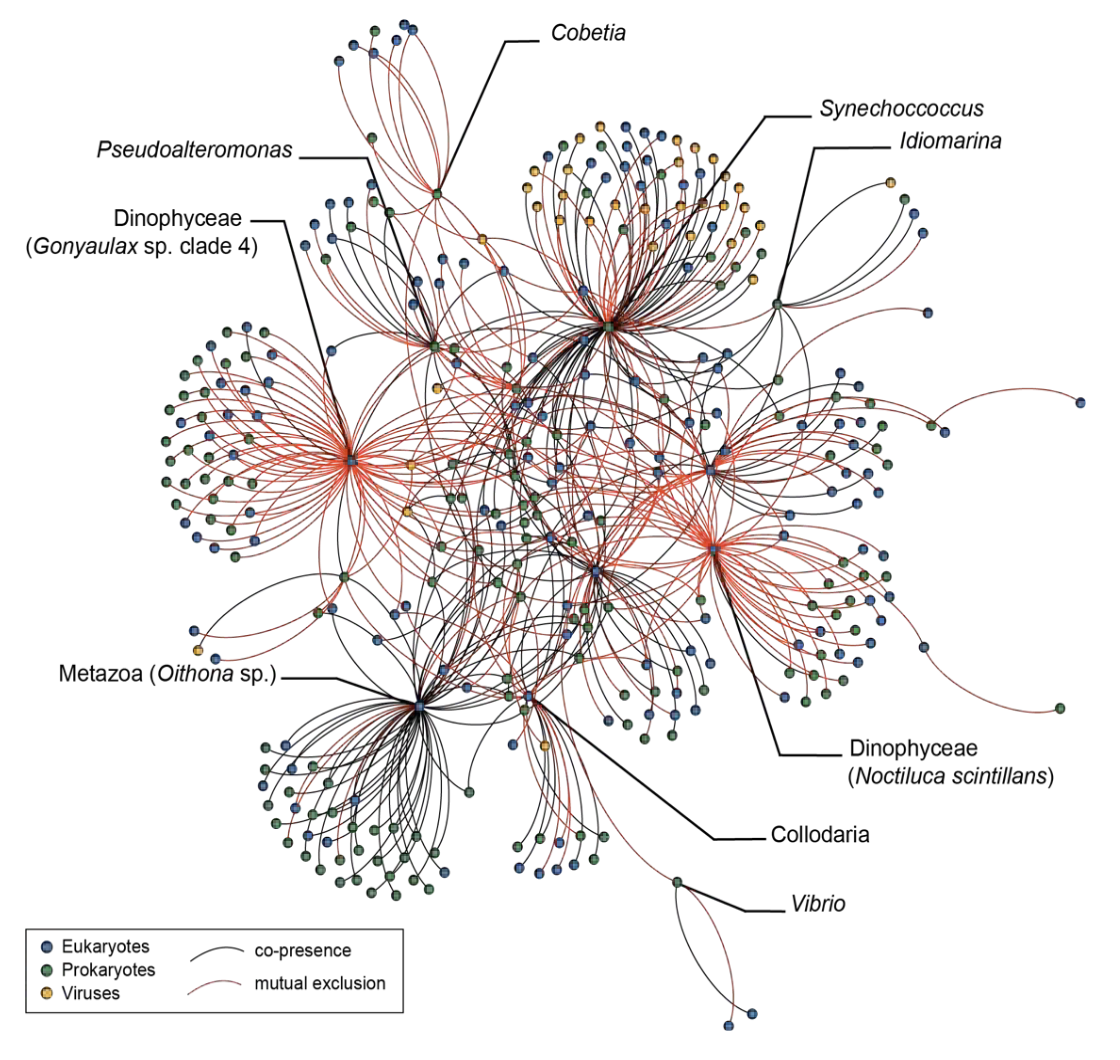
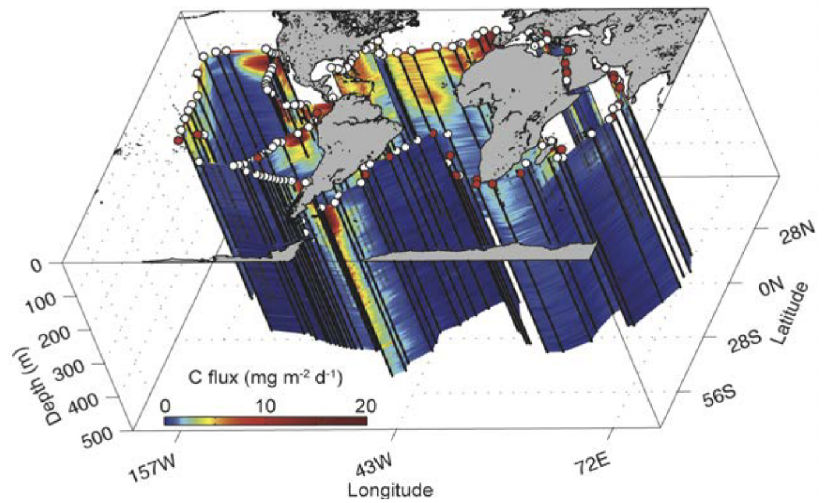
Figure 7. How Tara Oceans global eco-systemic data allow scaling up from genes or biomarkers’ communities to global ecosystem behavior. Left panel: Schematic representation of the biological carbon pump in the ocean. Atmospheric CO2 is transformed into organic carbon by phytoplankton in the sunlit layer, then incorporated into the highly-complex plankton interactome, and partly exported to deep oceanic layers and sediments through dead organisms, fecal pellets, and marine ‘snow’ (drawing from A. de Chastenet; note that depth, organisms and particle sizes are not to scale). Right side: we used systems-biology graph-based methods to integrate eukaryotic, prokaryotic and viral ‘omics’ data (top), and correlate their respective sub-networks to carbon export fluxes down to 500 m, as estimated at each Tara Oceans station from particle size distribution and abundance measured with the underwater vision profiler (UVP) (bottom). We could also identify keystone taxa or genes in the subnetworks (hubs) that can predict accurately carbon export.
